Original Research
SARS-CoV-2 Epitope Presentation by Class II HLA Genotypes Common in North American Populations: A Proposed Computational Approach for Vaccine Efficacy Evaluation
Laura Leclair1, Constantin Polychronakos2,3
Published online: September 2022
1Department of Microbiology and Immunology, McGill University
2Department of Pediatrics (cross-appointment in Human Genetics), McGill University
3Research Institute of the McGill University Health Centre
Corresponding Author: Laura Leclair, email laura.leclair@umontreal.ca
DOI: 10.26443/mjm.v21i1.907
Abstract
Background: Human Leukocyte Antigen (HLA) gene polymorphisms between ethnic groups have been shown to play a role in the heterogeneity of response to SARS-CoV-2, in terms of COVID-19 disease severity and susceptibility, in addition to socioeconomic factors. It was predicted that this finding may extend to vaccine responsiveness.
Purpose: To the best of our knowledge, this study was the first that aimed to predict and evaluate the effectiveness of four COVID-19 vaccines across North American ethnic groups, in terms of their ability to trigger CD4+ T cell help, based on class II HLA allele frequencies.
Methods: : Various databases including the Immune Epitope Database (IEDB) were used in this computational approach. The number of peptide-HLA high-affinity pairs between the most common HLA II haplotypes and SARS-CoV-2 peptides in various vaccine types were retrieved and compared between ethnicities. From this, the efficiency of antigen presentation to CD4+ T cells was evaluated, a crucial component in the context of vaccination for cellular immunity and support in antibody generation.
Results: Multiple discrepancies in vaccine effectiveness for ethnic minorities relative to the Caucasian group, overrepresented in vaccine clinical trials, were highlighted. Recommendations were issued in terms of which vaccine types could be most effective for particular ethnicities.
Conclusion: There exists a genetic basis for differential responses to vaccines among ethnic groups in North America. However, given the multifactorial nature of vaccine responsiveness and limitations of computational methods, this study offers future research directions to undertake before the findings can be transferred to clinical and public health settings.
Tags: HLA antigens, Vaccines, Ethnicity, COVID-19, SARS-CoV-2
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for causing Coronavirus disease 2019 (COVID-19) with infections ranging from asymptomatic to fatal. Structural proteins encoded within its genome are spike (S), envelope (E), membrane (M) and nucleocapsid (N). (1) SARS-CoV-2-specific antibodies, CD4+ T cells, and CD8+ T cells are at the core of the adaptive immune response against SARS-CoV-2. The focus of this study was on CD4+ T cells only, as they play a crucial role in natural infection and vaccination in terms of T cell help in antibody generation. They are also critical in maintaining long-term memory B cells and humoral immunity. (2)
The four main types of COVID-19 vaccines contain: whole virus (inactivated), the spike protein, the receptor-binding domain (RBD) of the spike protein, as well as spike and nucleocapsid proteins. (3-5) Most vaccine developers focus exclusively on the spike protein as it is the main target of neutralizing antibodies. (2) Yet, it has been shown that the generation of antibodies against spike relies on CD4+ T cells recognizing any component of the SARS-CoV-2 genome. (6) Grifoni et al. demonstrated via an ex vivo T cell assay that a SARS-CoV-2-specific CD4+ T cell response, correlated with antibody generation, may be triggered against almost all SARS-CoV-2 proteins. Thus, they recommended including more structural proteins in vaccines to better represent the infection observed in COVID-19. (7) This may amplify CD4+ T cell responses, in turn reinforcing humoral immunity through T cell help in generating antibodies.
CD4+ T cells recognize SARS-CoV-2 peptide-Human Leukocyte Antigen II (HLA II) complexes on the surface of antigen presenting cells to become activated and differentiate into their effector subsets. (8) The major HLA class II genes are HLA-DPA, HLA-DPB, HLA-DRA, HLA-DRB, HLA-DQA, and HLA-DQB. They encode highly polymorphic proteins, whose allele frequencies vary between ethnic groups, based on historical exposure to different pathogens. Each allele has varying binding affinity and can only bind a portion of the processed viral peptides, affecting the efficiency of antigen presentation. (9) Consequently, HLA alleles can either confer susceptibility or protection to infection or disease severity, as shown by epidemiological analyses and small-scale cohort studies of HLA typed individuals. (10-16) Heterogeneity in infection susceptibility and disease outcome can also be predicted by computational HLA and SARS-CoV-2 peptide association studies. Barquera et al. observed in a computational approach that the frequencies of strongest and weakest binders to various coronaviruses differ among populations from different geographic regions worldwide. (17) In the case of SARS-CoV-2, Copley et al. predicted similar protective CD4+ cellular immunity potential, based on HLA II haplotypes, across 25 human populations of different ethnicities in the United States. (18)
Other computational studies aiming to design the most effective multi-epitope COVID-19 vaccines predicted heterogeneity in vaccine response based on ethnicity. He et al. found that their designed vaccine may confer less protection for people of African descent based on HLA coverage. (19) Remarkably, Liu et al. designed vaccines predicted to have high coverage for Asian, Black, and White ancestry individuals. (20)
Ethnic minorities in North America are disproportionately affected by COVID-19, both in terms of risk of infection and mortality. (21) Socioeconomic factors have been extensively reported to explain such disparities, but the role of genetic factors remains understudied. (22-25) Further, participants from ethnic minorities are unjustifiably underrepresented in vaccine clinical trials. (26)
The rate of reported cases of COVID-19 on Indigenous reserves is 183% higher than in the general Canadian population. (27) Since geographically isolated Indigenous communities are not protected by the herd immunity of urban centers and may have a higher susceptibility to COVID-19, assessing vaccine effectiveness is critical.
Real-world, post-hoc vaccine effectiveness studies that have started to emerge mainly focus on the general population. While elderly or healthcare workers, for example, have been part of distinct subgroup analyses, ethnic groups have not. (28)
The underrepresentation of ethnic minorities in clinical trials may persist in vaccine effectiveness studies. Computational tools have proven extremely useful in SARS-CoV-2 vaccine development and may provide a solution by predicting vaccine effectiveness for special populations. (29)
A particularly relevant example is Liu et al.'s analysis of the Moderna, Pfizer-BioNTech, and AstraZeneca vaccines. They showed that African Americans and Asians could have a slightly increased risk of vaccine ineffectiveness in silico, but the findings remain to be confirmed clinically. (30)
Hence, evidence for the implication of HLA polymorphism and varying allele frequencies in COVID-19 heterogeneity is provided in the literature. This means different HLA haplotypes could also influence responsiveness to vaccines through presentation of different HLA II immunopeptidomes to CD4+ T cells. The purpose of this study is to predict and evaluate vaccine effectiveness in terms of the ability to trigger the CD4+ T cell help required in antibody generation, based on the HLA II haplotypes common in ethnically diverse North American populations.
Methods
Retrieving allele frequencies
The Allele Frequency Net Database (AFND) stores gene frequencies in the form of alleles, haplotypes, or genotypes from worldwide populations. (31) The "HLA classical allele frequency search" tool was used to retrieve the most common HLA II alleles in Canada and the United States (i.e., North American populations) for the HLA-DPA1, -DPB1, -DQA1, -DQB1, -DRB1 class II loci (Supplemental Material 1).
Grouping by principal component analysis (PCA) and computing weighted averages
To determine if the raw data for the individual populations in each ethnic group sample could be merged based on variance, PCAs were performed using R software 4.0.3 (R Core Team, Vienna, Austria) for each ethnic group separately. (32) From the resulting PCA biplots, pools or groups of populations were created based on the Euclidean distance in a plot of the first and the second component. The weighted averages of the allele frequencies based on sample sizes of the populations were computed for the various pools or groups. The weighted average frequencies of the most common alleles were summed to reach a minimum of 70% total coverage of the population, in line with the principle of herd immunity. In certain cases, multiple combinations were made, when alleles were particularly common. Finally, a merged list of alleles for all ethnic groups was created (Supplemental Material 2, 3).
Haplotype associations
HLA-DP and -DQ must be considered as haplotypes in the Immune Epitope Database (IEDB), while only the beta chain of HLA-DR alleles may be specified because the alpha chain is invariable. Thus, haplotypes were retrieved from AFND and the literature (Supplemental Material 1). (31, 33-39)
Retrieving COVID-19 vaccine contents and protein sequences
Four vaccine types (whole virus, spike, RBD and spike and nucleocapsid) were selected for analysis and their protein sequences were retrieved in FASTA format from the NCBI GenBank (Accession number: MN908947.3) (Supplemental Material 4). (40)
Using IEDB's MHC II binding prediction tool
Using the bioinformatics tool Split FASTA, (41) the various protein sequences were separated into peptides of 15 amino acids in length, with an overlap of 10 amino acids, to reduce redundancy in the results. (42) The protein sequences and the HLA haplotypes were inputted into IEDB's MHC II Binding Prediction Tool to predict peptide-HLA binding affinities and immunogenicity. (39, 42) The prediction method selected was "IEDB recommended 2.22" to ensure the best predictor or algorithm for each allele was used. To compare the binding affinities obtained from various algorithms, IEDB outputs a percentile rank for each peptide-HLA "hit." Lower percentile ranks suggest higher peptide-HLA affinity (Supplemental Material 4). (43)
Calculating weighted counts, total counts, and plotting count histograms
Weighted counts were computed by multiplying the number of peptide-HLA hits or score for each allele/haplotype by their frequency. Sums of the weighted counts for each ethnic group were then calculated, for each type of vaccine according to their protein contents, to arrive at total counts of the number of peptide-HLA hits for both top 1st and top 10th percentiles. The top 10th percentile data for all vaccine types except RBD (i.e., less relevant than other types) were distributed along percentile intervals. Count histograms were plotted from the sum of weighted counts along each interval (Supplemental Material 4).
This is a descriptive report that presented estimates of the HLA II and SARS-CoV-2 proteome affinities based on pre-existing data, without attempting to compare them. Thus, statistical analyses were not performed.
Results
Population definitions
HLA allele frequencies in population samples from all sources were subjected to PCA to determine how they could be pooled. It was not necessarily possible to pool together all samples from different sources named after the same ethnicity. For example, since the Indigenous Canada population samples appeared as two distinct clusters in the PCA biplot, they were separated into two groups, Indigenous Canada #1 and #2 (Table 1).
| Table 1: Total counts of peptide-Human Leukocyte Antigen (HLA) hits for all vaccine types | Whole virus | Spike | RBD | Spike and nucleocapsid | ||||
| Percentile | Top 10th | Top 1st | Top 10th | Top 1st | Top 10th | Top 1st | Top 10th | Top 1st | Indigenous Canada #1 | 113 | 6.10‡ | 15.6 | 0.980‡ | 1.95 | 0.00‡ | 19.2 | 0.980‡ |
| Indigenous Canada #2 | 142 | 14.3 | 19.1 | 1.95 | 3.80 | 0.697 | 21.7 | 2.09 |
| Indigenous US #1 | 95.2 | 5.29‡ | 10.6 | 0.315‡ | 1.78 | 0.00‡ | 13.3 | 0.426‡ |
| Indigenous US #2 | 177 | 18.7 | 22.8 | 2.97 | 5.24 | 0.779 | 25.5 | 2.97 |
| Indigenous US + Canada #1 | 177 | 18.6 | 22.7 | 2.97 | 5.23 | 0.778 | 25.5 | 2.97 |
| Indigenous US + Canada #2 | 95.2 | 5.29‡ | 10.6 | 0.315‡ | 1.78 | 0.00‡ | 13.3 | 0.426‡ |
| African American #1* | 63.9* | 5.55* | 8.06* | 0.635* | 1.59* | 0.173* | 9.59* | 0.668* |
| African American #2 | 137 | 10.7 | 14.7 | 0.579‡ | 2.34 | 0.544 | 17.1 | 0.886‡ |
| Caucasian† | 167† | 19.4† | 18.6† | 2.23† | 4.59† | 0.813† | 20.7† | 2.23† |
| Polynesian | 131 | 9.85 | 16.7 | 1.89 | 2.87 | 0.987 | 19.9 | 1.89 |
| Asian (General) | 111 | 9.14 | 16.2 | 1.28 | 1.80 | 0.102‡ | 19.5 | 1.46 |
| South Asian | 115 | 11.0 | 17.4 | 1.36 | 3.93 | 0.290 | 20.3 | 1.59 |
| East Asian | 109 | 9.42 | 15.9 | 1.34 | 2.88 | 0.417 | 18.2 | 1.66 |
| Southeast Asian #1 | 125 | 13.9 | 18.7 | 1.91 | 4.66 | 0.655 | 21.0 | 2.16 |
| Southeast Asian #2 | 130 | 11.8 | 16.4 | 1.05 | 2.20 | 0.766 | 18.8 | 1.05 |
| Hispanic #1 | 108 | 8.48‡ | 15.7 | 1.01 | 3.01 | 0.171 | 18.3 | 1.25 |
| Hispanic #2 | 96.4 | 5.32‡ | 10.7 | 0.302‡ | 1.72 | 0.00‡ | 13.5 | 0.411‡ |
| Mestizo #1 | 198 | 25.9 | 21.9 | 3.24 | 6.69 | 1.30 | 22.5 | 3.24 |
| Mestizo #2 | 113 | 8.69 | 16.2 | 1.07 | 3.10 | 0.149 | 18.9 | 1.27 |
| Mixed | 167 | 17.0 | 19.2 | 1.92 | 3.98 | 0.654 | 21.8 | 2.14 |
| Arab | 108 | 9.30 | 14.0 | 1.27 | 3.10 | 0.264 | 17.1 | 1.71 |
* Underestimate
† Reference group
‡ Discrepancy relative to Caucasian reference group (for top 1st percentile only)
Certain minority ethnic groups were predicted to have fewer peptide-HLA hits within the top 1st percentile
Results for the top 10th and top 1st percentile peptide-HLA hits were tabulated (Table 1). The reference group used to highlight major discrepancies were Caucasian, as it is the most studied ethnicity. As expected, it was one of the top scoring groups across all four vaccine types, showing high total counts of predicted peptide-HLA hits. Overall, the biggest discrepancies between ethnicities occurred within the top 1st percentile (i.e., the strongest peptide-HLA bindings). In fact, the bottom five scores for each type of vaccine were lower than the Caucasian score by at least a factor of 2, and up to a factor of 8 in the RBD vaccine.
An underestimate of the true total counts arose for African American #1 group because the coverage of the population only amounted to 40% instead of the set minimum of 70%, due to the unavailability of some data. Since African American #1 has a sample size of around 4,900 compared with approximately 480,000 for African American #2, the latter group was deemed more representative.
Increased counts of peptide-HLA hits among the lower percentiles may suggest better antigen presentation
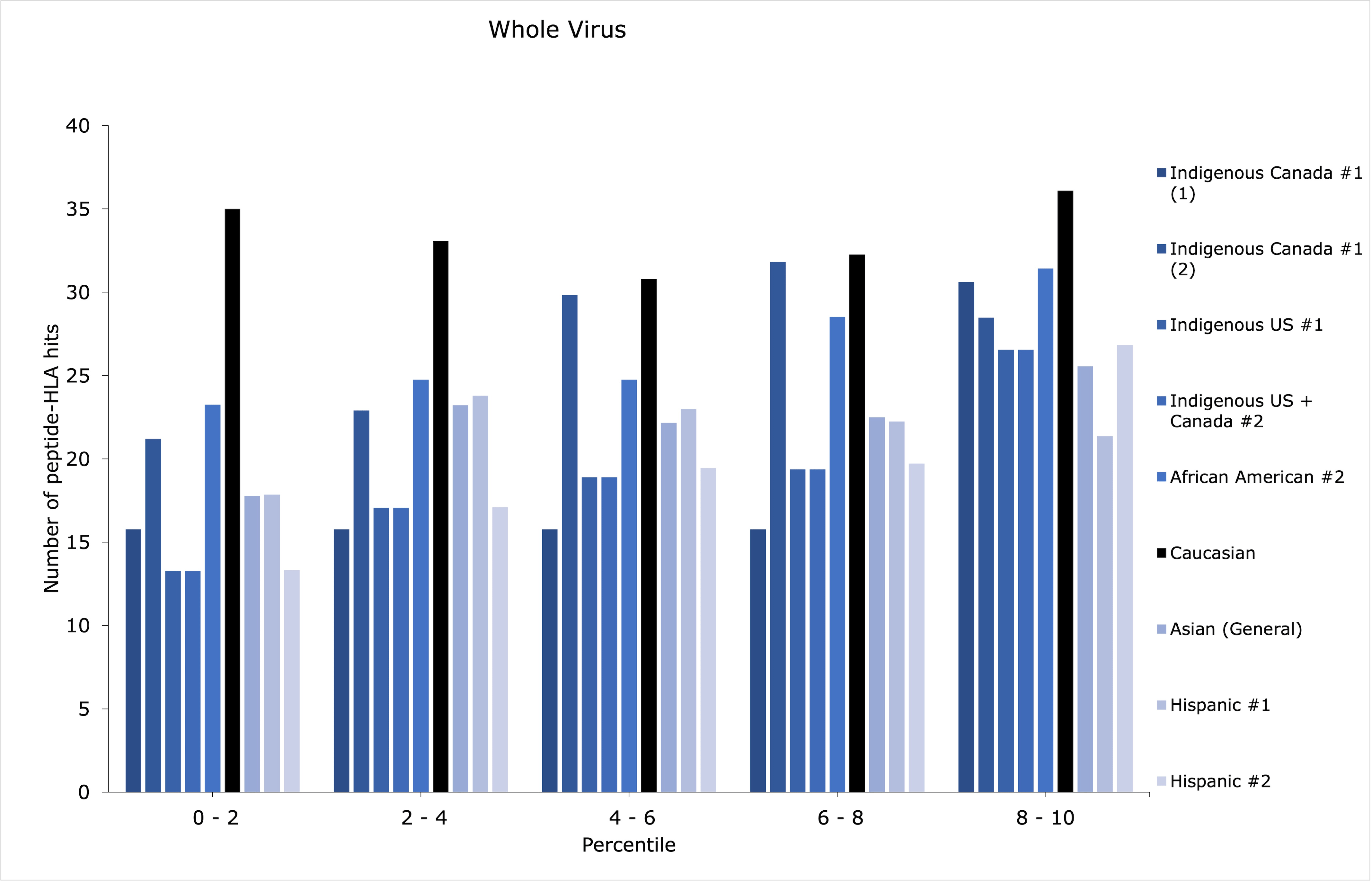
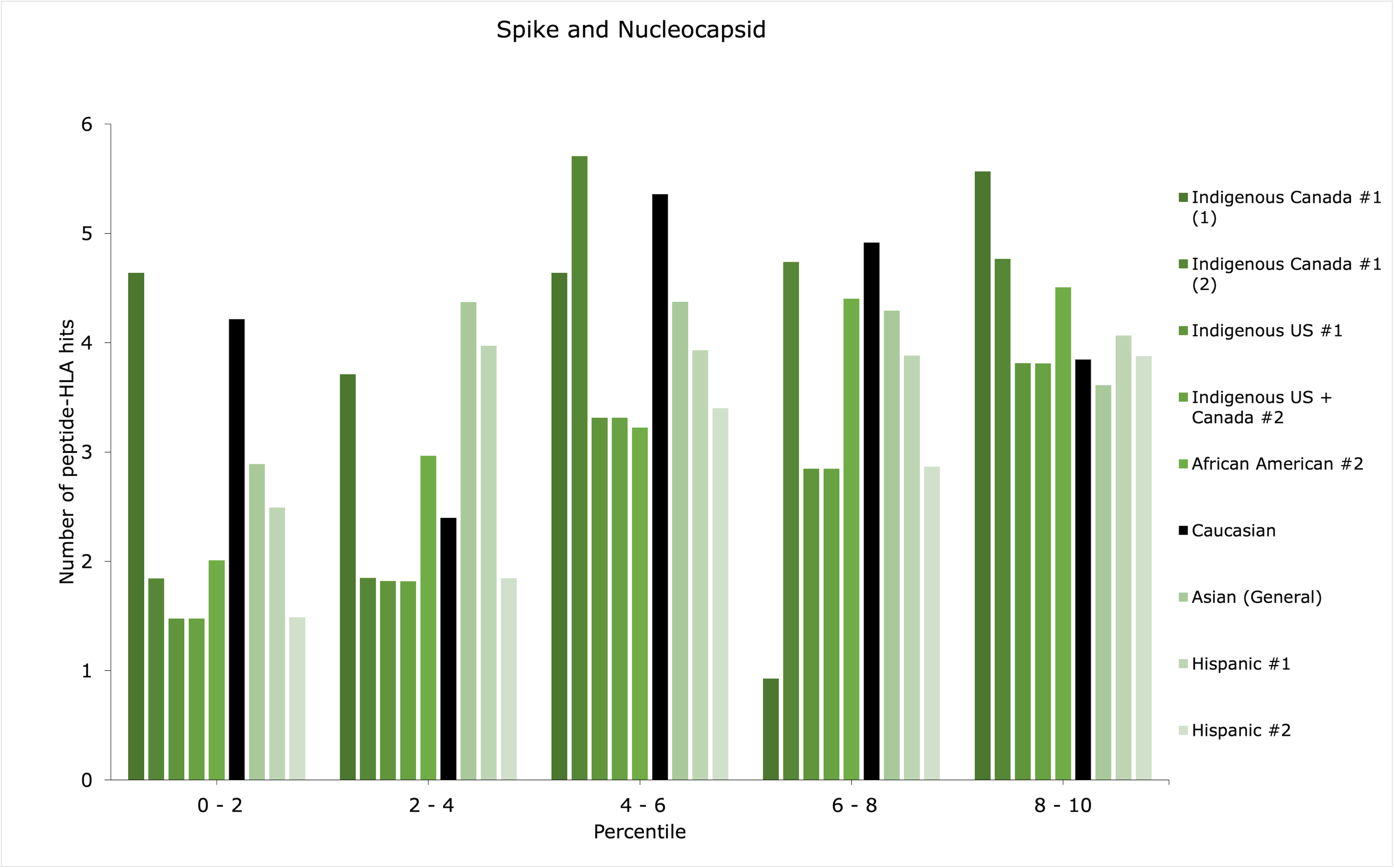
Although an ethnic group may have scored low within the top 1st percentile, it may have additional peptide-HLA hits with sufficient binding affinities for successful antigen presentation, for example, between the 1st and 5th percentiles. Thus, the distributions of peptide-HLA hits across the top 10th percentile were plotted in count histograms (Figures 1-3) for whole virus, spike, and spike and nucleocapsid vaccines, for the groups with the discrepancies identified in Table 1 (refer to Supplemental Material 4 for the rest of the data and RBD vaccine).
Patterns of distribution showing increased counts of peptide-HLA hits among the lower percentiles may suggest a greater amount of potentially stronger peptide-HLA hits, while increased counts among the higher percentiles (i.e., closer to the 10th percentile) could suggest a greater amount of weaker peptide-HLA hits. Most importantly, the former, most advantageous pattern of distribution may augment antigen presentation efficiency for the lowest-scoring groups in Table 1.
Caucasians showed high counts of peptide HLA-hits compared with ethnic minorities. This reflects that Caucasians could be, generally, more responsive to the four vaccine types
The four groups that showed major discrepancies across all vaccine types (Indigenous Canada #1, Indigenous US #1, Indigenous US + Canada #2, and Hispanic #2) could be particularly less responsive to RBD vaccines, as they showed a score of 0 for RBD in top 1st percentile (Table 1). Out of them, only allele combination #1 in the Indigenous Canada #1 group showed a pattern of distribution predicted to potentially augment the efficiency of antigen presentation for spike and spike and nucleocapsid vaccines, with slightly increased counts of low percentile values (Indigenous Canada #1 (1) in Figures 2 and 3). This could be indicative of enhanced effectiveness for two vaccine types for Indigenous peoples in Canada with the first combination of alleles. Overall, the type of vaccine to prioritize for the four low-scoring groups could be whole virus, for which the discrepancies in the top 1st percentile compared with Caucasian were less important than with other vaccine types (Table 1).
For African American #2, when looking at the spike as compared to spike and nucleocapsid vaccines, the addition of nucleocapsid enhanced the number of successfully predicted peptide-HLA hits as the gap with the Caucasian count was reduced. However, the top 10th percentile distributions were not deemed advantageous in providing additional successful peptide-HLA hits in both vaccine types (Figures 2 and 3). Instead, the difference in counts of peptide-HLA hits between African American #2 and Caucasian was less for the RBD vaccine than for whole virus. Thus, an RBD vaccine might provide stronger responses for African American #2 (Table 1).
For Asian (General) group, the opposite was predicted. The RBD only vaccine could be the least effective, and a more advantageous option could be the spike and nucleocapsid vaccine, providing less of a discrepancy with the Caucasian score (Table 1).
For Hispanic #1, whole virus was the lowest scoring vaccine. However, its top 10th percentile distribution seemed quite evenly distributed (Figure 1). This could suggest that strong peptide-HLA hits found among the lowest percentiles could provide enough response to the whole virus vaccine. The same cannot be said Hispanic #2, for which the spike and nucleocapsid vaccine may be preferable. Again, this can be explained by the smaller difference in counts between Caucasians and Hispanic #2 for this type of vaccine, compared with other types (Table 1).
In this computational approach to assess vaccine effectiveness across North American ethnic groups, it was proposed that differences in allele frequencies may result in different HLA II immunopeptidomes being presented to CD4+ T cells. In turn, this was thought to possibly translate to population-level differences in effective anti-SARS-CoV-2 immunity between ethnic groups, resulting in disparities in vaccine responsiveness. Barquera et al. compared the binding affinities in silico of HLA class I and II to SARS-CoV-2 peptides and found that the large majority do not bind HLA I molecules, whereas a proportion do bind HLA II molecules. (17) Furthermore, because CD4+ T cells are crucial in natural infection and vaccination for antibody generation, our COVID-19 vaccine effectiveness analysis focused solely on HLA II molecules. (2, 18) Complementary studies could investigate the role of HLA I.
Thus, vaccine effectiveness was evaluated in terms of counts and patterns of distribution of peptide-HLA hits, based on the interpretation that lower percentile peptide-HLA hits should provide the strongest bindings. More peptide-HLA hits within the top 1st percentile were predicted to offer better antigen presentation to CD4+ T cells, thereby improving effector response, support in antibody generation, and vaccine responsiveness. Less peptide-HLA hits within the top 1st percentile (or proportionally more hits within higher percentiles) were predicted to correlate with reduced vaccine responsiveness.
This hypothesis was based on an approach that we designed and proposed as a part of this manuscript and that has not yet been validated experimentally. The field of research on the immunological response to COVID-19 and its pathways is still evolving. The true implications of such predictions on antigen presentation and CD4+ cellular immunity cannot be adequately confirmed without in vitro and in vivo studies. Furthermore, although peptide-HLA hits are necessary for antigen presentation by HLA molecules, they may not be sufficient for T cell recognition. Nonetheless, Copley et al. validated their theoretical data by comparing to a dataset of experimentally determined T cell epitopes and demonstrating that their in silico approach did not overestimate the number of immunogenic epitopes. (18) The general method and prediction algorithm (NetMHCIIpan) used in latter study are similar to those in our present approach.
Regarding African American #2, He et al.'s multi-epitope vaccine design was also predicted to confer less protection for people of African descent based on HLA coverage, although the specific vaccine content differed from the ones tested here. (19)
While three Indigenous ethnic groups scored lower than Caucasians, the three other Indigenous groups (Indigenous Canada #2, Indigenous US #2 and Indigenous US + Canada #1) generally scored close to or higher than the Caucasian reference (Table 1). This finding is supported by Barquera et al., who showed that Indigenous peoples in America could have particularly protective HLA haplotypes in some instances and against a range of multiple viruses including coronaviruses. (17) One possible reason for having both low-scoring and high-scoring Indigenous groups could be due to the different degrees of admixture with ethnicities that are better equipped to respond to COVID-19 vaccines, like European descent admixture. Further studies analyzing genome-wide association studies (GWAS) data could confirm or refute this conjecture. Alternatively, natural selection may have favored the strongest HLA binders in those Indigenous groups following European colonization and the introduction of new infectious diseases. (17)
In opposition to Copley et al.'s conclusion that all ethnic groups have similar HLA II antigen presentation potential, (18) the findings presented here demonstrated variability in vaccine effectiveness based on different antigen presentation patterns for all vaccine types investigated. This discrepancy may result from differences in methodology. Copley et al. only used the NMDP/Be The Match registry to extract HLA haplotype data, while we combined multiple sources from the AFND as registries are notoriously incomplete for ethnic minorities. Further, they restricted their peptide-HLA binding affinities analysis to the NetMHCIIpan-4.0 algorithm and their results to a percentile rank threshold of ⋜2, whereas our analysis included but was not limited to the NetMHCIIpan-4.0 algorithm, and it covered results up to the top 10th percentile.
As previously suggested by Liu et al., African American and Asian populations, in general, scored lower than the Caucasian group when looking at the spike vaccine. In addition, the present study evaluated more vaccine types and a wider range of ethnic groups, including Indigenous populations, filling the gaps in Liu et al.'s work. (30)
Our results supported the recommendations by Zelba et al. and Grifoni et al. to include more proteins in vaccines than just spike or RBD. (6-7) Indeed, for certain ethnic groups and their characteristic HLA haplotypes, the whole virus, or the spike and nucleocapsid vaccines were predicted to be more effective.
In addition to the limitation related to the purely predictive basis of potential antigen presentation, before the recommendations issued from this evaluation can be transferred to the clinical and public health settings, the methodological limitations must also be acknowledged. Although computational methods provide rapid and cost-effective approaches, the prediction of peptide-HLA hits is an imperfect science due to the nature of predictive algorithms based solely on chemical structure and lack consideration for the actual in vivo environment in antigen presentation. In addition, populations were defined by visual interpretation of the various PCA pools, which would sometimes overlap and were not necessarily clearly defined. Also, HLA II binding predictions are known to be more challenging and less accurate than HLA class I, but their accuracy has greatly improved in the last decade. (42) Nonetheless, the limitation of using pre-existing HLA allele frequency data to predict viral infection outcomes could be alleviated by integrating HLA typing in vaccine clinical trials to clinically evaluate vaccine effectiveness for various HLA genotypes and ethnicities. A review paper by Sohail et al. on the performance of computational methods in the context of COVID-19 offered promising evidence of their practical significance but warned that improvements must be made before predictions can apply to real-world vaccine developments. (44)
Predictions may nonetheless impel researchers to validate such data with experimental and immunogenicity testing, and to undertake larger-scale epidemiological and clinical studies. Investigating new variants of SARS-CoV-2 in the approach used here could elucidate the consequences of mutations on HLA binding and thereby on vaccine effectiveness. Finally, the investigation of other biological factors such as the effects of polymorphisms in genes like ACE2 and TMPRSS2, (45) blood groups, hormonal balances, and co-morbidities, (46) in relation to ethnicity and vaccine effectiveness, are equally important perspectives for future research directions.
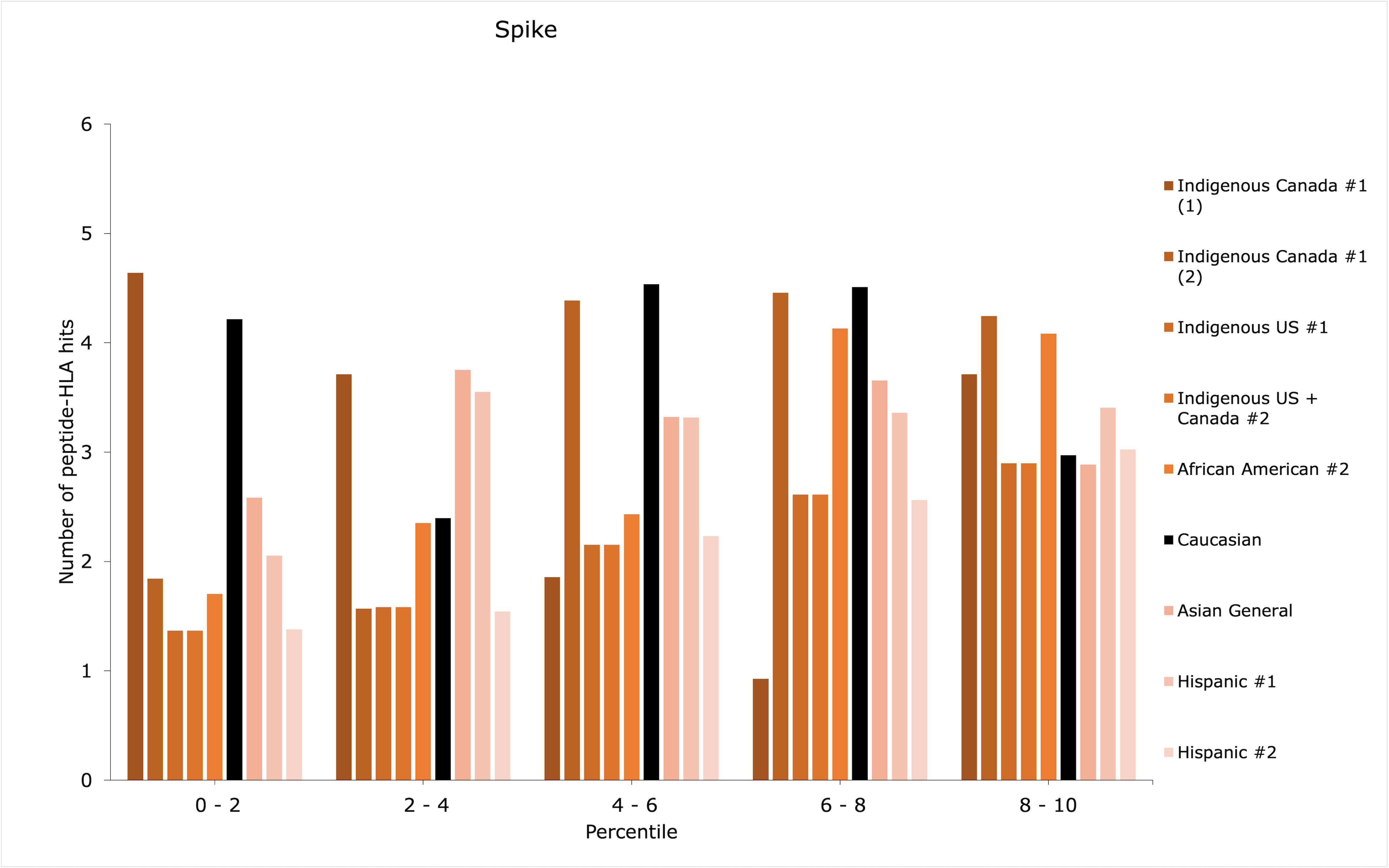
Discussion
Comparison of vaccine efficacy predictions with other studies
Methodological limitations and future research directions
References
Supplementary Material
Supplemental Material 1
HLA II Allele Frequencies for Canadian Populations of Various Sample Sizes
Supplemental Material 2
Sample PCA Code for Indigenous Canada
Supplemental Material 3
PCA Biplots for Ethnic Groups with 3 or More Variables
Supplemental Material 4
Haplotype Inputs for the Immune Epitope Database (IEDB) Analysis

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.